Assistant Professor
Degrees and Appointments
- Assistant Professor 2022-present, University of Arizona, Department of Chemistry & Biochemistry
- Assistant Professor 2017-2022, University of Wyoming, Department of Chemistry
- Postdoctoral Research Fellow, University of Cambridge, 2012 - 2017
- Ph.D. Organic Chemistry, University of Delaware, 2013
- B.S. in Biochemistry, with honors, Salisbury University, 2006
Awards and Honors
- 2022: University of Wyoming Presidential Scholarly Achievement Award
- 2021: NIH Outstanding Investigator Award (R35 MIRA)
- 2021: NSF CAREER award
- 2020: Thieme Chemistry Journals Award
- 2014–2016: Marie-Curie International Incoming Postdoctoral Fellowship
- 2012: Sigma-Aldrich Graduate Student Innovation Award
- 2012: Brennie A. Hackley Jr. Excellence in Research Award
- 2006-2007: Chemistry-Biology Interface Pre-Doctoral Training Program Fellowship
- 2006 American Institute of Chemists: Collegiate Achievement Award
Before joining UArizona CBC, Dr. Taylor held the postion of Assistant Professor in the Department of Chemistry at the University of Wyoming, from 2017 through August of 2022. Whilst at Wyoming, he developed a research program that is underpinned by an interest in developing organic transformations for use at the chemistry-biology interface. Michael has received an NSF CAREER award, a Thieme Chemistry Journals award, an NIH outstanding investigator award, and a Presidential Scholarly Achievement Award from the University of Wyoming.
Dr. Taylor's group will design, develop, and apply new organic chemical transformations that can be used to explore the chemistry-biology interface in living systems. Through these chemistries, which form non-natural covalent bonds with biomolecules, his group aims to study how biological systems work in real-time as well as how these systems change with respect to disease states or other perturbations. The results of these studies can have broad implications ranging from enhancing our basic understanding of biology to discovering new ligands and biomolecular targets for drug discovery. However, chemistries that are truly biocompatible must be remarkably robust in nature and thus their development presents a unique and exciting chemical challenge.
Research
Chemical transformations that create non-natural covalent bonds on biomolecules are truly empowering as they enable new approaches to the study of biological processes, drug discovery and therapeutic design, as well as the design of functional materials. However, reactions that modify biomolecules must meet a stringent set of requirements in order to function in complex biological environments and thus their development presents a unique and exciting chemical challenge.
The overarching research objectives in the Taylor group are to (1) design new and mechanistically unusual reactions that chemically modify biomolecules and (2) to apply them towards the study of how biological systems work and change dynamically in a native environment such as a living cell. Spanning the chemistry-biology interface in this fashion creates a highly multidisciplinary and collaborative environment where lines of inquiry merge diverse concepts ranging from physical organic chemistry, mechanistic photochemistry and synthesis to cellular/molecular biology and proteomics.
This multidisciplinary approach is represented in our recent work in harnessing photo-induced electron transfer (PET) as a mechanism for a new photochemically-driven protein ligation reaction. We demonstrated that this could be achieved by pairing the inherently photo- and redox-lability of Tryptophan (Trp) residues by with an N-carbamoyl pyridinium salt that simultaneously couples photo-induced electron transfer (PET) with Trp to a radical fragmentation/recombination process to carbamylate Trp residues site-selectively in high conversion and yields (Figure 1A. We also demonstrated that the photophysical and electrochemical properties of 1 can be readily tuned such that either [Trp]*-pyridinium PET (Trp is a photoreductant) or Trp->[pyridinium]* PET (Trp is a reductive quencher) can be invoked to drive the labelling chemistry.
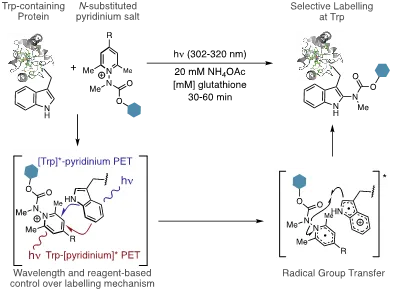
We have refined the pyridinium scaffold to include a donor-acceptor relationship between the two aryl groups (Figure 2). This relationship enables photoconjugation at Trp using visible light as a trigger, and at concentrations of protein and probe that are an order of magnitude more dilute than those used with our first-generation probes without sacrificing reaction times (10-60 min). These mild conditions enabled us to conduct global chemoproteomic profiling of Trp residues in live cell culture. Beyond demonstrating biocompatibility, this experiment enabled us directly label and identify >200 hyper-reactive Trp residues in their native environment. These types of data sets can then be used a basis for discovering “hotspots” that are of biochemical significance, as well to find new targets and sites for drug discovery.

Select Publications
1) Hoopes, C.R.‡; Garcia, F.G.‡; Sarkar, A.M.; Kuehl, N.J.; Barkan, D.T.; Collins, N.J.; Meister, G.E.; Bramhall, T.R.; Hsu, C-H.; Jones, M.D.; Schirle, M; Taylor, M.T. Donor-Acceptor Pyridinium Salts for Photo-Induced Electron Transfer Driven Protein Modification in Peptides, proteins and proteomes. J. Am. Chem. Soc. 2022, 144, 6227-6236. DOI: 10.1021/jacs.1c10536
‡Denotes equal contribution.
2) Taylor, M.T. Photochemical protein modification in complex biological environments: recent advances and considerations for future chemical methods development*. Biol. Chem. 2022. 403, 421-431. DOI: 10.1515/hsz-2021-0351.
3) Orellana, N.V.; Taylor, M.T. Targeting Tryptophan for Tagging Through Photo-Induced Electron Transfer. Synlett. 2021, 32, 1371-1378. DOI: 10.1055/a-1479-6366
4) Tower, S.J.; Hetcher, W.H.; Myers, T.E.; Kuehl, N.J.; Taylor, M.T. Selective Modification of Tryptophan Residues in Peptides and Proteins Using a Biomimetic Electron Transfer Process. J. Am. Chem. Soc. 2020, 142, 9112-9118. DOI: 10.1021/jacs.0c03039
5) Scinto, S.L.; Ekanayake, O.; Senevriatne, U.; Pigga, J.E., Boyd, S.J.; Taylor, M.T.; Liu, J.; am Ende, C.W; Rozovsky, S.; Fox, J.M. Dual-Reactivity trans-Cyclooctenol Probes for Sulfenylation in Live Cells Enable Temporal Control via Bioorthogonal Quenching. J. Am. Chem. Soc. 2019, 141, 10932-10937. DOI: 10.1021/jacs.9b01164
6) M.T. Taylor, J.E. Nelson, M.G. Suero, M.J. Gaunt A Protein Functionalization Platform Based on Selective Reactions at Methionine Residues.” Nature. 2018, 562, 563-568. DOI: 10.1038/s41586-018-0608-y
Complete List of Published Works:






