Professor of Chemistry and Biochemistry
Degrees and Appointments
- A.B. 1967-70, University of California at Santa Cruz
- Ph.D. 1970-75, University of California at Santa Cruz
- NIH Postdoctoral Fellow 1976-79, Biozentrum of the University of Basel (Switzerland)
- Research Scientist 1976-79, Max-Planck-Institute for Medical Research, Heidelberg (Germany)
- NIH Postdoctoral Fellow 1979, University of California at Berkeley
Awards and Honors
- Associate Editor, Biophysical Journal, 2016-present
- Avanti Award of the Biophysical Society, 2014
- Elected Fellow of the Biophysical Society, 2013
- Research Corporation Galileo Circle Fellow, 2013
- Elected Fellow of the American Association for the Advancement of Science (AAAS), 2012
- Elected Fellow of the American Physical Society, 2011
- Fellow of Japan Society for the Promotion of Science, 2003
- Senior Fulbright Fellow (University of Florence, Italy), 2000
- Röntgen-Professorship of Physics (Germany), 1999
- NIH Research Career Development Award, 1985-90
- Alfred P. Sloan Foundation Research Fellow, 1983-85
The Brown lab conducts research at the cutting edge of molecular spectroscopy and biophysical chemistry as well as nanoscience and naotechnology. We carry out fundamental research in molecular spectroscopy together with applications to membrane proteins, lipid bilayers, surfactants, and liquid crystals—soft matter in general. Our work spans fundamental science related to intermolecular interactions through to applied nanoengineering aspects, involving chemically tailored interfaces between lipids, proteins, and water. Novel experiments are put forth together with theory at the leading edge of macromolecular science, lipid nanotechnology, and (bio)physical chemistry. Applications encompass surface science, physical chemistry, condensed matter physics, materials science, and lipid nanotechnology. Our unique focus is on integrating structure, dynamics, and function of proteolipid and surfactant assemblies using NMR spectroscopy and related analytical and biophysical methods.
Our broad goal is to discover how molecular interactions lead to the emergence of material properties and biological function by combining molecular spectroscopy with dynamics simulations and experimental studies. In the area of molecular spectroscopy, we are applying nuclear magnetic resonance (NMR) technologies to liquid-crystalline materials and biomolecular systems. A major focus is the use of experimental and theoretical NMR methods to achieve new insights into functional molecular mechanisms. Indeed NMR is perhaps the most useful spectroscopic method in chemistry and biochemistry—it provides information about structure and dynamics with amazing breadth and versatility! Applications range from organic synthesis to protein structure elucidation, studies of membranes and (bio)polymers, and magnetic resonance imaging (MRI) of human subjects. Currently students are applying solid-state NMR spectroscopy to membrane liquid crystals and proteins using both solution and solid-state NMR methods. We also use solid-state NMR to investigate small molecule cofactors bound to membrane proteins. In the area of biophysical chemistry and nanoengineering, our laboratory is engaged in structure-function studies of proteolipid membranes. Chemical, physical, analytical, and biochemical methods provide a host of opportunities for cross-disciplinary training. Members of our interdisciplinary team conduct research at US National Laboratories (Oak Ridge, Brookhaven, SLAC), where X-rays and neutrons are used to investigate molecular structure and dynamics. Femtosecond X-ray nanocrystallography with a free electron laser is used to investigate conformational changes of membrane proteins. With this approach, we detect the "protein quake" underlying the large-scale conformational changes. Establishing how proteins like rhodopsin are affected by lipid bilayer deformation is another important emphasis of our highly motivated research team. Functional work uses rhodopsin as a canonical prototype for G-protein-coupled receptors (GPCRs), the targets of the majority of human pharmaceuticals worldwide. Notably we have designed lipid formulations using principles of nanoscience and technology that are capable of modulating the activity of membrane proteins like rhodopsin over a wide range.
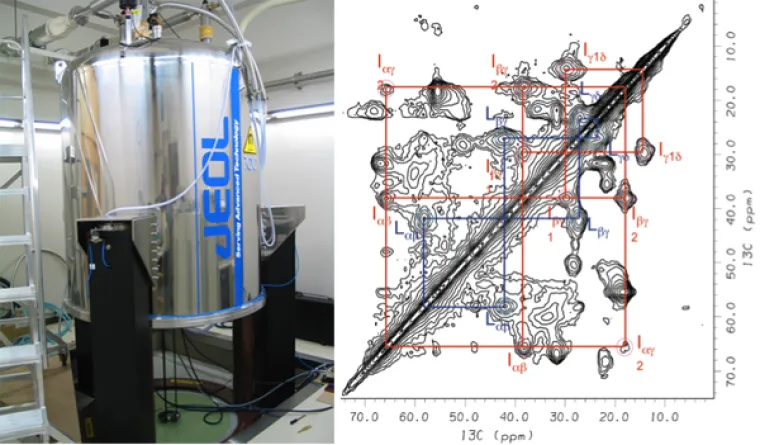
Solid-state NMR spectrometer with superconducting magnet and two-dimensional spectrum of a membrane protein. Note that the cross-peaks provide structural information.
Advancing the Frontiers of Biomolecular NMR Spectroscopy. Biomembranes are liquid-crystalline systems, and X-ray crystallography provides only a partial view of their functions. Our group has devised solid-state NMR methods (order parameter analysis, relaxation methods) in the first detailed studies of membrane structure and dynamics. We have investigated ion-induced changes in the head groups of membrane lipids; their interactions with various sterols; and the structural dynamics of membrane lipids and proteins. Our group achieved a breakthrough by introducing a mean-torque analysis of the order parameters of membrane liquid crystals. Additional structural work involves organic synthesis for isotopic labeling of the ligands for integral membrane proteins. Proteins are purified from natural sources or expressed in bacteria. Our group has studied membrane interactions with antimicrobial peptides and proteins, such as alpha-synuclein (implicated in neurodegenerative disorders), Ras (GTPase oncogene), and ATP-synthase (molecular motor). Currently my students apply solid-state NMR to study light induced changes of rhodopsin (visual receptor), and an amino acid transport protein from chloroplasts. We are also using these concepts to illuminate the roles of omega-3 polyunsaturated lipids in membranes. Additional new directions include NMR studies of medically important proteins involved with human health and disease.
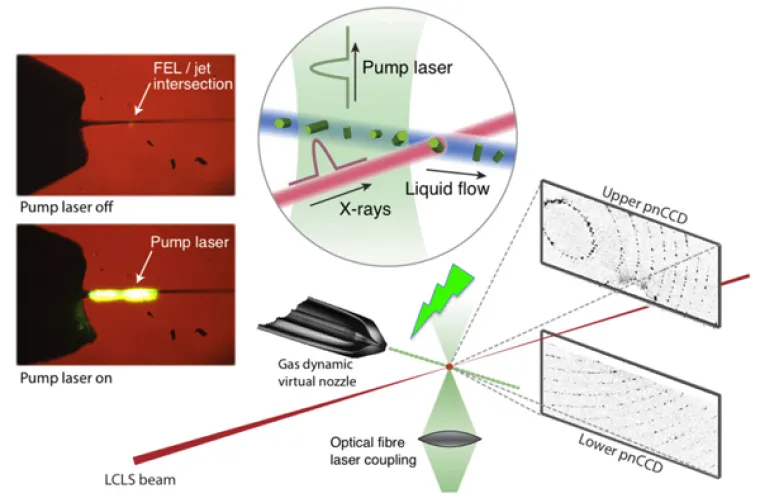
Laser excitation scheme used for pump-probe studies with femtosecond X-ray nanocrystallography of membrane proteins.
Femtosecond Nanocrystallography of Membrane Proteins. The advent of X-ray free-electron lasers (XFELs) opens up entirely new horizons in chemistry, biology, and physics. The XFEL pulses have a peak brilliance that is 109 stronger than any synchrotron X-ray radiation and enable time-dependent studies of protein dynamics to be conducted. Our team is currently investigating rhodopsin in nanocrystals using the short intense X-ray pulses of the free-electron laser at the Linac Coherent Light Source (LCLS) at SLAC National Accelerator Laboratory. Pump-probe studies of light activation are carried out in the sub-picosecond to microsecond range, and directly examine the fastest structural events of vision. Soon, we plan to extend the work to the millisecond time range, to probe the changes of rhodopsin that yield interaction with the G-protein, transducin. Current projects involve preparing nano/microcrystals of rhodopsin for pump-probe femtosecond X-ray studies. Additional time-resolved, wide-angle X-ray scattering (WAXS) studies of rhodopsin in detergent solutions investigate light-induced conformational changes ("protein quake"). The ultimate outcome will be a molecular movie of the activation process as a prototype for the Rhodopsin Family of G-protein-coupled receptors (GPCRs).
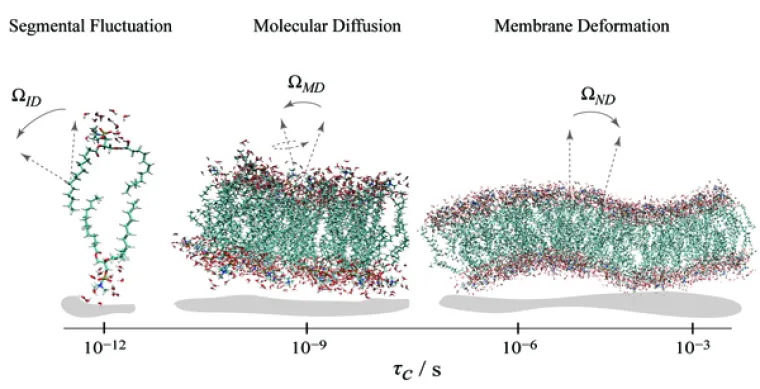
Solid-state NMR relaxation reveals phospholipid membrane dynamics over a range of time and length scales.
Relaxation Experiments and Theory Development in NMR Spectroscopy. Nuclear magnetic resonance (NMR) methods are among the premier experimental tools for obtaining knowledge of dynamics. Our experimental measurements of the magnetic field (frequency) dependence of the NMR relaxation rates are crucial for molecular dynamics (MD) simulations. Before we began our work in this area, interpreting the NMR relaxation times of biomolecules was viewed as too complicated to be solved in a meaningful way. However, we were able to achieve a solution with clear biophysical significance by combining experiment with theory. For lipid bilayers, the new model relates the energy landscape of the molecular fluctuations to the emergence of elastic properties over short distances, approaching the molecular dimensions. Knowing such properties allows a holistic interpretation of membrane structure and dynamics for understanding lipid interactions with cholesterol, peptides, and proteins. Projects include NMR relaxation studies of membrane lipid deformation by osmotic stress. The magnetic field (frequency) dependence of the relaxation is measured to identify the time and length scales of collective interactions. Additional projects involve cholesterol influences in lipid rafts, as well as lipid interactions with membrane peptides and proteins, and NMR relaxation studies of protein dynamics.
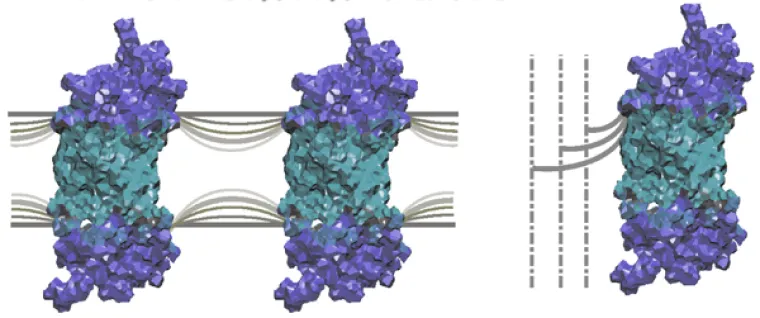
Flexible surface model (FSM) describes how conformational changes of membrane proteins depend on spontaneous curvature of the lipid bilayer.
Biophysics and Biochemistry of Membranes. Our studies of lipid-protein interactions have significantly advanced the field of biomembranes. Notably, we have extended the standard model (the fluid mosaic model) found in most biochemistry texts. We discovered how membrane lipids govern the conformational energetics of proteins by our innovation of a flexible surface model (FSM). In our picture, frustration (elastic deformation) of the membrane is counterbalanced by the hydrophobic mismatch of the lipids to the protein surface. Our bold new ideas explain how the tightly regulated lipids affect biological functions of proteins in signal transduction, ion conductance, and metabolite transport. Conformational changes involve elastic remodeling of the lipid bilayer that affects lipid-protein and lipid-peptide interactions (described by differential geometry). Allosteric regulation of membrane proteins is linked to influences of bilayer thickness, nonlamellar-forming lipids, detergents, and osmotic stress that are explained by the FSM as a new paradigm. Currently my students are investigating how properties such as curvature stress and membrane surface potential govern light activation of rhodopsin. The role of the membrane lipid composition in controlling activation of the G-protein (transducin) is also under study. Investigations of the roles of omega-3 polyunsaturated lipids in visual function and dysfunction are carried out. Our work illuminates how membrane properties underlie key cellular processes involved with signaling, ion channels, and drug transport.
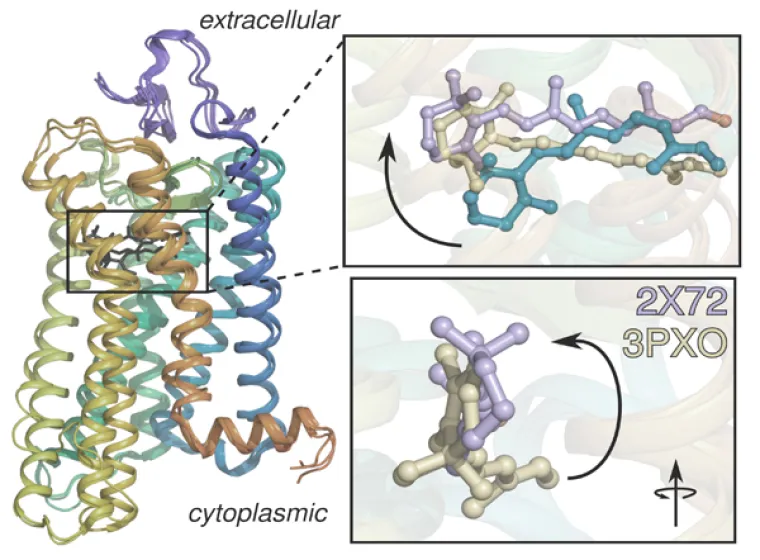
Three-dimensional structure of rhodopsin showing how retinal is more elongated in the active structures (purple and tan) than in the dark structure (cyan).
Discovering New Insights into Visual Signaling by Rhodopsin. Knowing how rhodopsin is activated by light is important for understanding retinal proteins and can contribute added insight into membrane mechanisms. Prior to our work in this area, rhodopsin was regarded as a simple on-off switch. Our team determined the solid-state NMR structure of the retinal ligand of rhodopsin, and the changes triggered by light. Surprisingly, we discovered that activation goes beyond a two-state conformational transition. To explain our findings, we introduced an ensemble activation model. We have proposed that the retinal cofactor initiates helix fluctuations on the microsecond-to-millisecond timescale in visual signaling. Our innovations help to reveal how mutations of rhodopsin are implicated in human diseases. Currently, we are combining solid-state NMR, electronic (UV-visible), and Fourier transform infrared (FTIR) spectroscopy to investigate how retinal triggers rhodopsin activation in membranes. By combining spectroscopic methods with functional assays, we aim to achieve a comprehensive understanding of rhodopsin activation as a prototype for various membrane proteins.
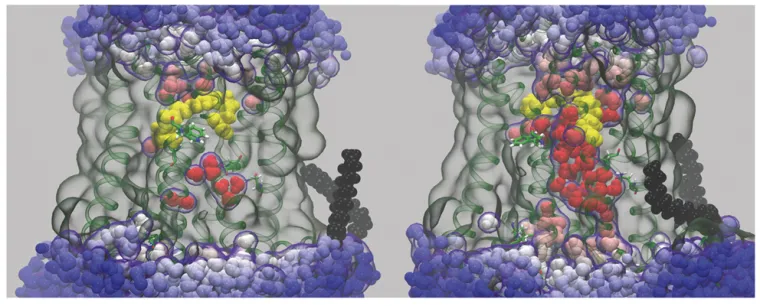
Molecular dynamics (MD) simulation showing an Influx of bulk water into rhodopsin following light-induced isomerization of the retinal cofactor
Research
Unraveling the Molecular Dynamics of G-Protein–Coupled Receptors. Rhodopsin is the prototype for the Family A G-protein–coupled receptors (GPCRs), which are the targets of the majority of human pharmaceuticals. When we began our work, the only X-ray structure for a GPCR was rhodopsin in the dark state. Our group has been combining molecular dynamics (MD) simulations with experimental solid-state NMR data to investigate the triggering of visual signaling by rhodopsin. All-atom MD simulations are conducted using supercomputers and graphics processor units (GPUs) to model rhodopsin activation, and its interactions with signaling proteins like transducin. We were first to discover a surprising influx of bulk water into the protein core giving activation of rhodopsin. Currently our research team is testing the relations between hydration, binding affinity, and transducin activation in visual signaling. Our molecular dynamics simulations and solid-state NMR studies point towards enhanced ligand flexibility, which explains the different retinal orientations seen in X-ray structures of active rhodopsin.
CHECK OUT THE BROWN LAB PAGE FOR THE LATEST NEWS!
VISIT ALSO THE UA CHEMICAL PHYSICS PROGRAM!!
Click on the links below to download pdf files of some of our latest publications:
- Cholesterol modulates membrane elasticity via unified biophysical laws. Kumarage, T. et al. Nat Commun 16, 7024 (2025). https://doi.org/10.1038/s41467-025-62106-0
- Flexible Surface Model for Lipid-Protein Interactions. Fried, S. D. E. & Brown, M. F. in Membrane Shape and Biological Function (CRC Press, 2025). https://www.taylorfrancis.com/chapters/edit/10.1201/9781003287650-2/flexible-surface-model-lipid-protein-interactions-steven-fried-michael-brown
- Pump-probe TR-WAXS Distinguishes Activation from Heating Effects in Ultrafast Rhodopsin Dynamics. Karpos, K. et al. SPring-8/SACLA Research Report 13, 175–182 (2025). https://doi.org/10.18957/rr.13.3.175
- Osmotic stress studies of G-protein-coupled receptor rhodopsin activation; Andrey V. Struts, Alexander V. Barmasov, Steven D.E. Fried, Kushani S.K. Hewage, Suchithranga M.D.C. Perera, Michael F. Brown, Biophys. Chem., 304, (2024), 107112, ISSN 0301-4622, https://doi.org/10.1016/j.bpc.2023.107112
- Efficient calculation of orientation-dependent lipid dynamics from membrane simulations; Milka Doktorova, George Khelashvili, Michael F. Brown, bioRxiv 2023.05.23.542012; doi: https://doi.org/10.1101/2023.05.23.542012
- Hidden water’s influence on rhodopsin activation. Bachler, Z. T. & Brown, M. F. Biophys. J. 123, 4167–4179 (2024). https://doi.org/10.1016/j.bpj.2024.11.012
- Biophysics of Membrane Stiffening by Cholesterol and Phosphatidylinositol 4,5-bisphosphate (PIP2)2023 ;Doole, F.T., Gupta, S., Kumarage, T., Ashkar, R., Brown, M.F. (2023). Biophysics of Membrane Stffening by Cholesterol and Phosphatidylinositol 4,5-bisphosphate (PIP2). In: Dantsker, A.R. (eds) Cholesterol and PI(4,5)P2 in Vital Biological Functions. Advances in Experimental Medicine and Biology, 1422. Springer, Cham. https://doi.org/10.1007/978-3-031-21547-6_2
- Molecular simulations and NMR reveal how lipid fluctuations effect membrane mechanics2023 ;Milka Doktorova, George Khelashvili, Rana Ashkar and Michael F. Brown, (2023) Bio Phys. J. 122, 984-1002, https://doi.org/10.1016/j.bpj.2022.12.007
- Cholesterol Stiffening of Lipid Membranes2022 ;Fathima T Doole, Teshani Kumarage, Rana Ashkar, Michael F Brown, J Membr Biol. 2022, 255(4-5):385-405. doi: https://doi.org/10.1007/s00232-022-00263-9.
- Phospholipid headgroups govern area per lipid and emergent elastic properties of bilayers2022 ;Trivikram R. Molugu, Robin L. Thurmond, Todd M. Alam, Theodore P. Trouard, and Michael F. Brown (2022) Bio phys J. 121, 4205-4220, https://doi.org/10.1016/j.bpj.2022.09.005
- Hydration-mediated G-protein–coupled receptor activation2022 ;Steven D. E. Fried , Kushani S. K. Hewage, Anna R. Eitel, Andrey V. Strutsa, Nipuna Weerasinghe,Suchithranga M. D. C. Perera, and Michael F. Brown (2022) Proc. Natl. Acad. Sci. U.S.A, 119, e2117349119, https://doi.org/10.1073/pnas.2117349119
- Cholesterol stiening of lipid membranes and drug interactions: Insights from neutron spin echo and deuterium NMR spectroscopy2022 ;Chapter 29 Sudipta Gupta. Fathima T. Doole, Teshani Kumarage, Milka Doktorova, George Khelashvili, Rana Ashkar, Michael F. Brown (2022), Academic Press, Cholesterol From Chemistry and Biophysics to the Clinic, 771-796, https://doi.org/10.1016/B978-0-323-85857-1.00037-7
- Ashkar, R., Doktorova, M., Heberle, F. A., Scott, H. L., Barrera, F., Katsaras, J., Khelashvili G., and Brown, M. F. (2021), The Universal Stiffening Effects of Cholesterol in Lipid Membranes. Proc. Natl. Acad. Sci. U.S.A., in press (invited).
- Fried,S. D. E., Lewis, J. W., Szundi, I.; Martinez-Mayorga, K., Mahalingam, M., Vogel, R, Kliger, D. S., and Brown, M. F (2021), Membrane Curvature Revisited—the Archetype of Rhodopsin Studied by Time-Resolved Electronic Spectroscopy, Biophys. J. 120, 440–452.
- Chawla, U.; Perera, S. M. D. C.; Fried, S. D. E.; Eitel, A. R.; Mertz, B.; Weerasinghe, N.; Pitman, M. C.; Struts, A. V.; and Brown, M. F. (2021), Activation of the G-Protein-Coupled Receptor Rhodopsin by Water, Angew. Chem. Int. Ed. 60, 2288–2295.
- Chakraborty, S., Doktorova, M., Molugu, T. R., Heberle, F., Scott, H. L. Dzikovski, B., Nagao, M., Stingaciu, L.-R., Standaert, R. F., Barrera, F., Katsaras, J., Khelashvili, G., Brown, M. F.*, and Ashkar, R.* (2020) *Co-corresponding author, How Cholesterol Stiffens Unsaturated Lipid Membranes, Proc. Natl. Acad. Sci. U.S.A. 117, 21896-21905.
- Mallikarjunaiah, K. J., Kinnun, J. J., Petrache, H. I., and Brown, M. F. (2019), Flexible Lipid Nanomaterials Studied by NMR Spectroscopy, Phys. Chem. Chem. Phys. 21, 18422–18457.
- Navidi, M., Yadava, S., Struts, A. V., Brown, M. F., Nesnas, N. (2018), Synthesis of 9-CD3-9-cis-Retinal Cofactor of Isorhodopsin, Tetrahedron Lett. 59, 4521–4524.
- Perera, S. M. D. C., Chawla, U., Shrestha, U. R., Bhowmik, D., Struts, A. V., Qian, S., Chu, X.-Q, and Brown, M. F. (2018), Small-Angle Neutron Scattering Reveals Energy Landscape For Rhodopsin Photoactivation, J. Phys. Chem. Lett. 9, 7064-7071.
- Brown, M. F. (2017), Soft Matter in Lipid-Protein Interactions, Annu. Rev. Biophys. 46, 379-410.
- Molugu, T. R., Lee, S., and Brown, M. F. (2017), Concepts and Methods of Deuterium NMR Spectroscopy Applied to Biomembranes, Chem. Rev. 117, 12087−12132.
- Perera, S. M. D. C., Chawla, U., and Brown, M. F. (2016), Powdered G-Protein–Coupled Receptors, J. Phys. Chem. Lett. 7, 4230-4235.
- Shrestha, U. R., Perera, S. M. D. C., Bhowmik, D., Chawla, U., Mamontov, E., Brown, M. F., and Chu, X.-Q. (2016), Quasi-Elastic Neutron Scattering Reveals Ligand-Induced Protein Dynamics of a G-Protein–Coupled Receptor, J. Phys. Chem. Lett. 7, 4130−4136.
- Chawla, U., Jiang, Y., Zheng, W., Kuang, L., Perera, S. M. D. C., Brown, M. F., and Liang, H. (2016), A Usual G-Protein-Coupled Receptor in Unusual Membranes, Angew. Chem. Int. Ed. 54, 1–6.
- Ashkar, R., Doktorova, M., Heberle, F. A., Scott, H. L., Barrera, F., Katsaras, J., Khelashvili G., and Brown, M. F. (2021), The Universal Stiffening Effects of Cholesterol in Lipid Membranes. Proc. Natl. Acad. Sci. U.S.A., in press (invited).
- Fried,S. D. E., Lewis, J. W., Szundi, I.; Martinez-Mayorga, K., Mahalingam, M., Vogel, R, Kliger, D. S., and Brown, M. F (2021), Membrane Curvature Revisited—the Archetype of Rhodopsin Studied by Time-Resolved Electronic Spectroscopy, Biophys. J. 120, 440–452.
- Chawla, U.; Perera, S. M. D. C.; Fried, S. D. E.; Eitel, A. R.; Mertz, B.; Weerasinghe, N.; Pitman, M. C.; Struts, A. V.; and Brown, M. F. (2021), Activation of the G-Protein-Coupled Receptor Rhodopsin by Water, Angew. Chem. Int. Ed. 60, 2288–2295.
- Chakraborty, S., Doktorova, M., Molugu, T. R., Heberle, F., Scott, H. L. Dzikovski, B., Nagao, M., Stingaciu, L.-R., Standaert, R. F., Barrera, F., Katsaras, J., Khelashvili, G., Brown, M. F.*, and Ashkar, R.* (2020) *Co-corresponding author, How Cholesterol Stiffens Unsaturated Lipid Membranes, Proc. Natl. Acad. Sci. U.S.A. 117, 21896-21905.
- Mallikarjunaiah, K. J., Kinnun, J. J., Petrache, H. I., and Brown, M. F. (2019), Flexible Lipid Nanomaterials Studied by NMR Spectroscopy, Phys. Chem. Chem. Phys. 21, 18422–18457.
- Navidi, M., Yadava, S., Struts, A. V., Brown, M. F., Nesnas, N. (2018), Synthesis of 9-CD3-9-cis-Retinal Cofactor of Isorhodopsin, Tetrahedron Lett. 59, 4521–4524.
- Perera, S. M. D. C., Chawla, U., Shrestha, U. R., Bhowmik, D., Struts, A. V., Qian, S., Chu, X.-Q, and Brown, M. F. (2018), Small-Angle Neutron Scattering Reveals Energy Landscape For Rhodopsin Photoactivation, J. Phys. Chem. Lett. 9, 7064-7071.
- Brown, M. F. (2017), Soft Matter in Lipid-Protein Interactions, Annu. Rev. Biophys. 46, 379-410.
- Molugu, T. R., Lee, S., and Brown, M. F. (2017), Concepts and Methods of Deuterium NMR Spectroscopy Applied to Biomembranes, Chem. Rev. 117, 12087−12132.
- Perera, S. M. D. C., Chawla, U., and Brown, M. F. (2016), Powdered G-Protein–Coupled Receptors, J. Phys. Chem. Lett. 7, 4230-4235.
- Shrestha, U. R., Perera, S. M. D. C., Bhowmik, D., Chawla, U., Mamontov, E., Brown, M. F., and Chu, X.-Q. (2016), Quasi-Elastic Neutron Scattering Reveals Ligand-Induced Protein Dynamics of a G-Protein–Coupled Receptor, J. Phys. Chem. Lett. 7, 4130−4136.
- Chawla, U., Jiang, Y., Zheng, W., Kuang, L., Perera, S. M. D. C., Brown, M. F., and Liang, H. (2016), A Usual G-Protein-Coupled Receptor in Unusual Membranes, Angew. Chem. Int. Ed. 54, 1–6.
- Kinnun, J. J., Mallikarjunaiah, K. J., Petrache, H. I., and Brown, M. F. (2015), Elastic Deformation and Area Per Lipid of Membranes: Atomistic View From Solid-State Deuterium NMR Spectroscopy, Biochim. Biophys. Acta 1848, 246-259.
- Struts, A. V., Chawla, U., Perera, S. M. D. C., and Brown, M. F. (2015), Investigation of Rhodopsin Dynamics in its Signaling State by Solid-State Deuterium NMR Spectroscopy, in Methods in Molecular Biology, Jastrzebska, B. (Ed.), Springer, pp. 133–158.
- Leftin, A., Molugu, T. R., Job, C., Beyer, K., Brown, M. F. (2014), Area per Lipid and Cholesterol Interactions in Membranes from Separated Local-Field 13C NMR Spectroscopy, Biophys. J. 107, 2274-2286.
- Leioatts, N., Mertz, B., Martinez-Mayorga, K., Romo, T. D., Pitman, M. C., Feller, S. E., Grossfield, A., and Brown, M. F. (2014), Retinal Ligand Mobility Explains Internal Hydration and Reconciles Active Rhodopsin Structures, Biochemistry 53, 376-385.
- Xu, X., Struts, A. V., and Brown, M. F. (2014), Generalized Model-Free Analysis of Nuclear Spin Relaxation Experiments, eMagRes 3, 275-286.
- Struts, A. V., and Brown, M. F. (2014), Structural Dynamics of Retinal in Rhodopsin Activation Viewed by Solid-State 2H NMR Spectroscopy, in Advances in Biological Solid-State NMR: Proteins and Membrane-Active Peptides, Separovic, F., and Naito, A. (Eds.), The Royal Society of Chemistry, Cambridge, pp. 320-352.
- Zhu, S., Brown, M. F., Feller, S. E. (2013), Retinal Conformation Governs pKa of Protonated Schiff Base in Rhodopsin Activation, J. Am. Chem. Soc. 135, 9391-9398.
- Leftin, A., Job, C., Beyer, K., and Brown, M. F. (2013), Solid-state 13C NMR Reveals Annealing of Raft-Like Membranes Containing Cholesterol by the Intrinsically Disordered Protein α-Synuclein, J. Mol. Biol. 425, 2973-2987.
- Kinnun, J. J., Leftin, A., and Brown, M. F. (2013), Solid-State NMR Spectroscopy for the Undergraduate Physical Chemistry Laboratory,J. Chem. Ed. 90, 123-128.
- Brown, M. F. (2012), Curvature Forces in Membrane Lipid-Protein Interactions, Biochemistry 51, 9782-9795.
- Struts, A. V., Salgado, G. F. J., Martinez-Mayorga, K., and Brown, M. F. (2011), Retinal Dynamics Underlie its Switch from Inverse Agonist to Agonist During Rhodopsin Activation, Nature Struct. Mol. Biol. 18, 392-394.
- Struts, A. V., Salgado, G. F. J., and Brown, M. F. (2011), Solid-State 2H NMR Relaxation Illuminates Functional Dynamics of Retinal Cofactor in Membrane Activation of Rhodopsin, Proc. Natl. Acad. Sci. U.S.A. 108, 8263-8268.
- Leftin, A., and Brown, M. F. (2011), An NMR Data Base for Simulations of Membrane Dynamics, Biochim. Biophys. Acta 1808, 818-839.
Nuclear Magnetic Resonance (NMR) Spectroscopy Lipid Nanoengineering & Material Science Lipid Biomembranes Biophysical Chemistry Protein Dynamics Liquid Crystals & Surfactants Soft Matter Membrane Biophysics Molecular Dynamics (MD) Computer Simulations Physical Chemistry






