Regents Professor Emeritus - Retired
Degrees and Appointments
- B.A. 1962, St. Olaf College
- A.M. 1964, Harvard University
- Ph.D. 1966, Harvard University
- University of Arizona,, Assistant Professor of Chemistry, 1968-72
- Associate Professor,1972-77, Professor, 1977-98
- Regents Professor, 1998-2006
- Regents Professor Emeritus, 2006 - present
Fields of Study: Biochemistry, Inorganic Chemistry
Research Specialties: Bioinorganic, Protein and Membrane Biochemistry, Spectroscopy/Molecular Structure
Awards and Honors
- College of Science Distinguished Career Teaching Award, 2005
- University of Arizona Graduate College and Professional Education Teaching and Mentoring Award, 2002
- Mortar Board Citation Award, University of Arizona Mortar Board National Senior Honor Society, 2002
- El Paso Natural Gas Foundation Faculty Achievement Award, 1994
- Senior Alexander von Humboldt Award, 1981, Reinvitation, 1992
- Fulbright Senior Scholar Award, 1989
- American Association for the Advancement of Science Fellow, 1971
Research
1. Molybdenum-containing Enzymes
Molybdenum is an essential trace element for all forms of life and over 50 molybdoenzymes are known that catalyze oxidation-reduction reactions involved in the metabolism of carbon, nitrogen and sulfur. The enzyme sulfite oxidase (SO) is required for normal neurological development in children, and the crystal structure of chicken liver SO from protein prepared in our group has provided a basis for interpreting fatal point mutations in the highly homologous human enzyme. In addition, the novel structure of the molybdenum center of SO provides a target for the synthesis of new active site models as well as a framework for interpreting spectra from the enzyme. The large separation (32 Å) between the molybdenum center and the b-type heme center of SO has raised fundamental questions about the process and pathways of intramolecular electron transfer in the protein. Our research has involved an integrated program of chemical, biochemical and biophysical studies that include: synthesis of new compounds, X-ray structure determination, theoretical calculations, kinetics, and many types of spectroscopy (CW- and pulsed EPR, electronic, magnetic circular dichroism (MCD), resonance Raman, NMR, K- and L-edge X-ray absorption, and photoelectron (PES)) (see publications).
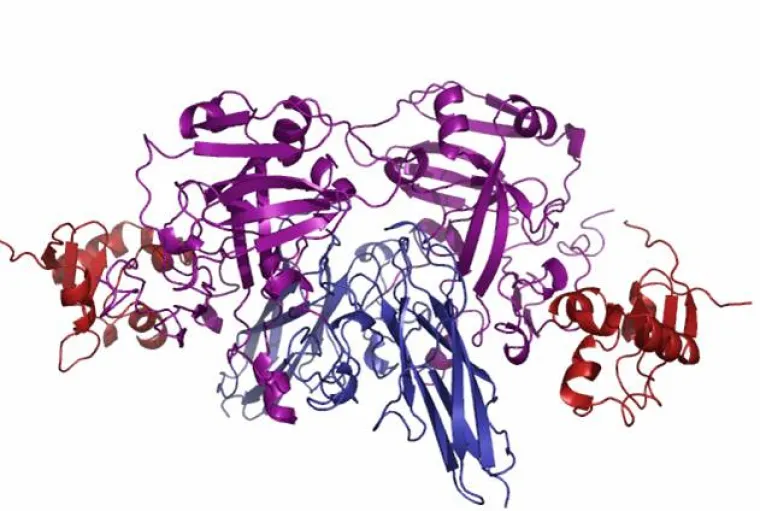
Structure of Chicken Sulfite Oxidase
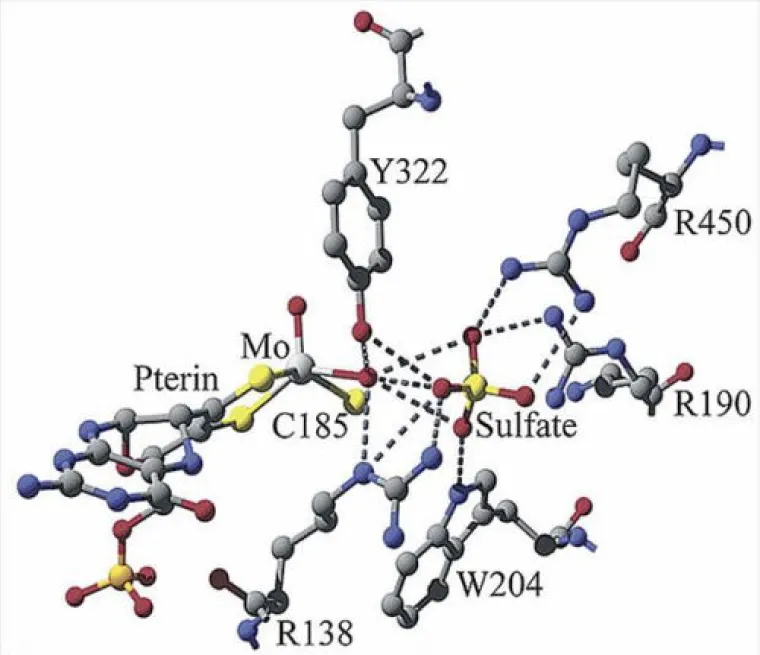
Sulfite Oxidase Active Site
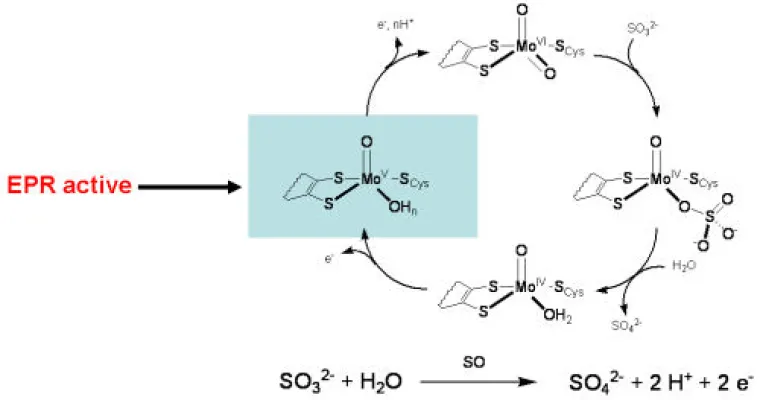
Sulfite Oxidase Proposed Mechanism
2. Variable Frequency Pulsed Electron Paramagnetic Resonance (EPR) Spectroscopy
The variable frequency pulsed EPR capabilities at the University of Arizona have enabled us to directly probe the structures of the transient Mo(V) states that are intermediates in the catalytic cycles of molybdenum enzymes. From Electron Spin Echo Envelope Modulation (ESEEM) and pulsed Electron-Nuclear Double Resonance (ENDOR) methods we have determined the active site structures of SO and DMSO reductase, and the effects of point mutations on these structures. Advances in pulsed EPR instrumentation in the EPR Facility enabled us to track oxygen atoms in the catalytic cycle through couplings of Mo(V) to 17-O and 33-S in isotopically enriched frozen solutions. A worldwide network of collaborators has provided access to a wide range of molybdenum enzymes and their mutants. The goal of these collaborative studies has been to obtain key insight about the catalytic mechanisms of molybdenum enzymes, while simultaneously developing new pulsed EPR methodologies for studying metalloproteins.
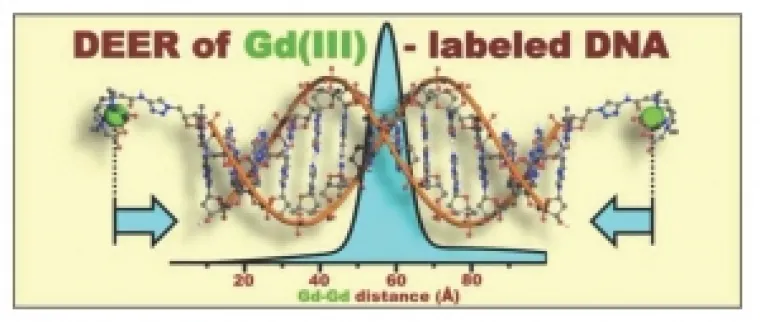
3. Measurement of Long Distances in Macrobiomolecules
Double electron-electron resonance (DEER) spectroscopy and Gd(III) spin-labels have been developed in collaboration with Arnold Raitsimring of the EPR Facility, Professor D. Goldfarb of the Weizmann Institute and Professor T. Meade of Northwestern University, to measure long distances (up to 100 Ångstroms) in macrobiomolecules. Shown below is an initial example of distance determination of ~60 Å in a 14-mer of DNA (Journal of Magnetic Resonance 210 [2011] 59–68).
Selected publications, for a complete list see the Enemark Group page.
Enemark, J. H. “{Moco}n, (n = 0–8): A General Formalism for Describing the Highly Covalent Molybdenum Cofactor of Sulfite Oxidase and Related Mo Enzymes.” J. Inorg. Biochem. 2022, 231, 111801 (https://doi.org/10.1016/j.jinorgbio.2022.111801). Invited contribution to memorial issue dedicated to Prof. R. H. Holm; cover feature.
Yang, J.; Enemark, J. H.; Kirk, M. L. “Metal-Dithiolene Bonding Contributions to Pyranopterin Molybdenum Enzyme Reactivity.” Inorganics, 2020, 8(3), 19 (https://doi.org/10.3390/inorganics8030019).
Stein, B. W.; Yang, J.; Mtei, R.; Wiebelhaus, N. J.; Kersi, D. K.; LePluart, J.; Lichtenberger, D. L.; Enemark, J. H.; Kirk, M. L. “Vibrational Control of Covalency Effects Related to the Active Sites of Molybdenum Enzymes.” J. Am. Chem. Soc. 2018, 140, 14777−14788 (DOI: 10.1021/jacs.8b08254).
Enemark, J. H. “Consensus Structures of the Mo(V) Sites of Sulfite-Oxidizing Enzymes Derived from Variable Frequency Pulsed EPR Spectroscopy, Isotopic Labelling and DFT Calculations.” Dalton Trans. 2017, 46, 13202-13210 (DOI: 10.1039/c7dt01731f). Invited contribution to themed issue on Inorganic Spectroscopy.
Kappler, U.; Enemark, J. H. “Sulfite Oxidizing Enzymes.” J. Biol. Inorg. Chem. 2015, 20, 253-264 (DOI 10.1007/s00775-014-1197-3).






